SARS-CoV-2 spike protein persists in the skull-meninges-brain axis in COVID-19 patients
Spike protein is sufficient to induce brain pathological and behavioral changes in mice
Spike protein enhances brain vulnerability and exacerbates neurological damage in mice
mRNA vaccines reduce, but do not eliminate, the spike burden
SARS-CoV-2 infection is associated with long-lasting neurological symptoms, although the underlying mechanisms remain unclear. Using optical clearing and imaging, we observed the accumulation of SARS-CoV-2 spike protein in the skull-meninges-brain axis of human COVID-19 patients, persisting long after viral clearance. Further, biomarkers of neurodegeneration were elevated in the cerebrospinal fluid from long COVID patients, and proteomic analysis of human skull, meninges, and brain samples revealed dysregulated inflammatory pathways and neurodegeneration-associated changes. Similar distribution patterns of the spike protein were observed in SARS-CoV-2-infected mice. Injection of spike protein alone was sufficient to induce neuroinflammation, proteome changes in the skull-meninges-brain axis, anxiety-like behavior, and exacerbated outcomes in mouse models of stroke and traumatic brain injury. Vaccination reduced but did not eliminate spike protein accumulation after infection in mice. Our findings suggest persistent spike protein at the brain borders may contribute to lasting neurological sequelae of COVID-19.
SARS-CoV-2 infection is associated with numerous neurological complications1,2,3 and a heightened risk of ischemic strokes.4,5 Even mild cases of COVID-19 can result in long-term effects, including brain fogging and reduced gray matter thickness.6,7,8 While some studies detected SARS-CoV-2 in brain tissue,3,9,10,11,12 others did not,12,13,14,15,16 possibly due to differences in patient populations and technical issues.17 Nevertheless, even without detectable virus RNA in the brain parenchyma, signs of widespread immune activation were detected.18 The lack of convincing evidence for the presence of whole viruses and especially viral replication in the brain led to the hypothesis that virus-shed proteins circulating in the bloodstream may promote an inflammatory response independent of direct viral infection of the affected organs, including the brain.19,20 Indeed, the spike protein, one of the critical structural proteins of SARS-CoV-2 mediating cell entry, has been shown to affect endothelial function in vitro21,22,23 and in vivo24,25 inducing Toll-like receptor (TLR) 2 or TLR4-mediated inflammatory responses after systemic injection in mice.26,27 Spike protein also directly binds to fibrin, triggering the formation of proinflammatory blood clots that cause systemic thromboinflammation and neuropathology.28 Long-lasting persistence of the spike protein in humans has been detected in immune cells (at least 15 months)29 and in the blood plasma (at least 12 months).30 However, the distribution of the spike protein in different organs, tissues, or regions of the body and its functional implications remain largely unclear.31,32
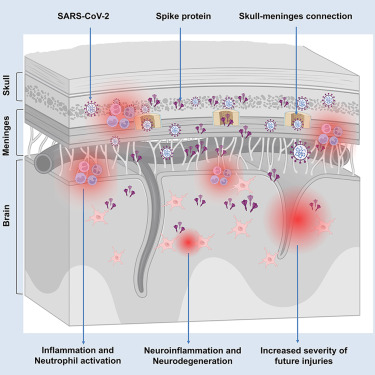
Here, we used advanced optical tissue clearing technologies33,34,35,36,37,38 to identify tissues targeted by SARS-CoV-2 in large human samples and across entire mouse bodies. We demonstrated spike protein distribution and accumulation across various tissues in both mice and post-mortem COVID-19 patient samples, including in the skull-meninges-brain axis, an anatomical structure challenging to preserve in its entirety. Using mass spectrometry-based proteomics, we found dysregulated neutrophil and inflammatory pathways in both mouse and human samples. Notably, the spike protein was found in the recently discovered skull-meninges connections (SMCs), where it might contribute to the COVID-19-induced inflammatory response, aligning with their role in the interactions of the peripheral immune system and the brain.34,39,40,41,42,43 Lingering spike protein was detected in the skulls of patients who had recovered from COVID-19 but died of non-COVID-19 causes, and biomarkers of neurodegeneration were elevated in the cerebrospinal fluid (CSF) of individuals with long COVID symptoms.
In healthy mice, spike protein injected into the skull marrow induced neuronal stress and inflammation in the brain parenchyma, potentially through activation of the mitogen-activated protein kinase (MAPK)-c-Jun N-terminal kinase (JNK) signaling pathway. Peripherally administered spike protein also triggered neuroinflammation and anxiety-like behavior. Additionally, spike exposure exacerbated the effects of cerebral ischemia and traumatic brain injury (TBI). We further demonstrated that the Pfizer/BioNTech vaccine reduced spike protein levels in the brain and brain borders of mice infected with the Omicron variant of SARS-CoV-2. These findings suggest that SARS-CoV-2 and its spike protein accumulating at brain borders may contribute to both immediate and long-term neurological effects.
To investigate the potential involvement of the brain border regions and the recently discovered SMCs34,39,40,44 in COVID-19, we used our small-micelle-mediated human organ efficient clearing and labeling (SHANEL) human tissue clearing method.36 Thereby, we analyzed the distribution of SARS-CoV-2 in post-mortem skull tissues (parietal bone next to sagittal suture) with the attached dura mater from patients who died of acute COVID-19 (Figure 1A; Table S1).
Figure 1 SARS-CoV-2 spike protein persistency in patient post-mortem skull and brain tissues
(A) Sampling site in the human skull with meninges.
(B) Representative images of spike protein antibody and propidium iodide (PI) labeling in post-mortem patient skull with meninges. The dura mater (blue) is segmented manually based on autofluorescence.
(C) Representative confocal images of spike and nucleocapsid protein in human skull marrow and meninges.
(D) Representative image of spike protein and collagen IV antibody staining in COVID-19 patient brain with meninges. Yellow arrowheads indicate spike protein is not colocalized with blood vessels.
(E) Confocal images of spike protein antibody labeling in the COVID-19 patient’s meninges and brain cortex. Yellow arrowheads indicate the spike protein in close proximity to the neuron’s nucleus.
(F) Summary of quantitative reverse-transcription PCR (RT-qPCR) and immunostaining results in skull, meninges, and brain samples. Each column represents one patient (Table S1).
(G) Representative images of human skull samples with spike protein antibody staining. Samples from patient that died before the COVID-19 pandemic were used as control.
(H) Single-molecule array (SIMOA) neurology assay of CSF samples. Upper row: all samples; lower row: only patients > 50 years. The graph displays mean ± SEM. One-way ANOVA with Fisher’s least significant difference (LSD) test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, nonsignificant.
See also Figure S1 and Videos S1 and S2.
We identified the spike and nucleocapsid protein in the skull marrow niches, SMCs, and meninges of these patients (Figures 1B, 1C, and S1A; Video S1). In the skull marrow, 45% of the spike protein was found outside the blood vessels (Figure S1B), suggesting that spike protein extravasated into the skull marrow tissue. We found 27% of spike protein colocalized with the Iba1+ myeloid cells, and 16% Iba1+ cells colocalized with spike protein in the skull marrow (Figure S1C).
Section view of a COVID-19 patient’s skull sample shows spike protein in the skull marrow, with the background in gray and the spike protein in red. A 3D reconstruction of a COVID-19 patient’s skull with dura sample reveals spike protein in the skull marrow, skull-meninges connections, and meninges. The background is blue, the spike protein is red, nuclei labeled with PI are cyan, and the dura mater is yellow.
Similarly, we detected spike protein in the brain frontal cortex (adjacent to the skull) of post-mortem COVID-19 patients (Figure 1D; Video S2). High-resolution confocal imaging showed spike protein in the perinuclear space of meningeal cells and near NeuN-positive neurons in the cortex (Figure 1E). Additionally, spike protein colocalized with tyrosine hydroxylase (TH)-positive neurons (Figure S1D). On average, 2.3% of the skull marrow cells, 2.8% of meningeal cells, and 1% of brain tissue cells were spike protein positive (samples from four patients, Figure S1E). Myeloid cell activation was observed in COVID-19 brain tissue (Figure S1F), with a moderate correlation between spike protein presence and Iba1 signal intensity (Figure S1G). Consistent with the propensity of COVID-19 patients to develop hemorrhage in the brain parenchyma,45 we detected microbleeds in both spike protein-positive and protein-negative brain regions (Figure S1H).
3D reconstruction of COVID-19 patient brain with meninges sample. Section view shows spike protein in meninges and brain vasculature. The background is shown in gray, the spike protein in red, and Iba1 labeling of myeloid cells in cyan.
SARS-CoV-2 RNA was detected in 8/16 skull samples and 6/12 meninges samples, while we detected the spike protein in 13/16 skull samples and 9/12 meninges samples, including all PCR-positive samples (Figure 1F). By contrast, all brain cortex samples we inspected were PCR negative, but we detected spike protein in 8/16 and nucleocapsid protein in 3/16 of these brain samples (Figure 1F). The presence of spike protein in the absence of viral load in the brain and meninges might suggest either a specific uptake mechanism to the brain or a longer half-life of spike protein compared with SARS-CoV-2 viral particles and active viral replication.
In Germany, 46% of the population has reportedly been infected with SARS-CoV-2.46 We anticipated that sampling skulls from individuals who died of non-COVID-19-related causes during the pandemic might include those who previously had COVID-19, allowing us to detect persistent spike protein in the skull marrow niches.
We examined skulls of patients who died from non-COVID-19-related causes in 2021 and 2022 and identified spike protein in 10 out of 34 samples (Figures 1G and S1I; Table S2), indicating that spike protein can persist long after infection resolution, potentially contributing to long-term COVID-19 symptoms. Notably, a correlation has been observed between persistent circulating spike protein in plasma and post-acute sequelae of COVID-19.30
To better understand the relationship between COVID-19 and neurological symptoms, we examined patient CSF and found elevated levels of Tau in long COVID patients compared with recovered patients and non-COVID-19 controls (Figure 1H; Table S3). Notably, Tau levels were also increased in acute COVID-19 patients, suggesting that neurodegeneration may be linked to both acute and long-term symptoms. Additionally, neurofilament light chain (NfL) and glial fibrillary acidic protein (GFAP) levels were significantly higher in long COVID patients than in recovered patients, suggesting ongoing neurodegeneration (Figure 1H). Although spike protein persists in solid tissues, its levels were below the assay’s detection limit and were not detected in patient CSF, likely due to the high rate of fluid exchange in the CSF.47
To determine the full tissue tropism of SARS-CoV-2, we used tissue clearing and imaging of whole mouse bodies with a GFP-expressing version of the virus.48 We measured virus distribution 5 days post-infection (dpi) in K18-hACE2 mice (transgenic mice expressing human angiotensin-converting enzyme 2 [hACE2] by the human keratin 18 promoter)49 (Figure 2A) and detected GFP and spike protein in various organs, including the lung, liver, intestine, and brain (Figures 2B and 2C), consistent with findings in COVID-19 patients.10,50,51 We also examined viral persistence in mice at 28 dpi after recovery from acute symptoms (Figure 2D). While GFP was absent from the skull marrow and brain cortex, spike protein was still detectable in the head at 28 dpi (Figures 2E and S2A–S2C).
Figure 2 Visualization of SARS-CoV-2 viral tropism in whole mouse bodies and SARS-CoV-2 spike protein persistency in skull marrow and brain
(A–E) SARS-CoV-2-GFP aerosol infection in K18-hACE2 mice. (A) Illustration of SARS-CoV-2-GFP infection and imaging of whole mouse body. (B) 3D reconstruction of whole mouse body showing localization of virus-encoded GFP and viral spike protein. (C) Representative optical sections of the GFP and spike signal at 5 dpi. (D) Representative images of spike protein in the whole mouse body at 5 and 28 dpi. (E) Representative images in the skull marrow and brain at 5 and 28 dpi.
(F–H) Alexa-647 conjugated protein intravenous injection in WT mice. (F) 3D reconstruction of whole mouse body 30 min after injection, showing the localization of WT spike protein (not binding mouse ACE2), N501Y spike S1 protein (binding mouse ACE2), and influenza hemagglutinin (HA). (G) Individual optical sections. The white box area was zoomed in for a detailed assessment of the brain borders. (H) Representative images of main internal organs with spike S1 (N501Y) protein and dextran-labeled vasculature. Arrowheads indicate regions with spike S1 protein.
See also Figure S2 and Video S3.
We also examined the distribution of a fluorescently labeled recombinant spike S1 protein carrying the N501Y mutation, which can bind to wild-type (WT) mouse ACE2 and is present in many SARS-CoV-2 variants.52,53 At 30 min post-intravenous injection into WT mice, spike S1 was found to localize near blood vessels in most organs, including the heart, lung, liver, kidney, intestine, thymus, spleen, pancreas, and brain (Figures 2F–2H, S2D, and S2E; Video S3). By contrast, the influenza H1N1 surface protein hemagglutinin (HA) was primarily detected in the liver and lung (Figure 2F). WT spike protein (from the original SARS-CoV-2 strain Wuhan-Hu-1) was used as a non-ACE2 binding control, and it predominantly accumulated in the liver (Figure 2G).
3D reconstruction of an intact mouse torso scanned by light-sheet microscopy. The background is represented in gray, the spike S1 protein in yellow glowing color. Spike S1 protein in various organs are visualized in high contrast over the background. High-resolution visualization of spike S1 protein (green) and dextran-labeled vessels (magenta) in mouse organs, captured using light-sheet microscopy. Section views include the liver, intestine, heart, spleen, and pancreas.
This multi-organ distribution aligns with the broad expression pattern of ACE2.54,55 Staining of the blood-brain barrier (BBB) revealed that spike S1 crossed the BBB, as previously suggested.56 However, spike protein mainly colocalized with pericytes (PDGFR+) and not with neurons (β-tubulin III+) (Figure S2F).
Substantial accumulation of spike S1 was also observed in the skull marrow niches and SMCs (Figure S2G). Spike S1 was found in the marrow of the tibia and femur, indicating its capacity to reach bone marrow niches (Figure S2H), and the localization of spike protein in bone marrow and meninges may correlate with ACE2 expression in these compartments.57,58
We noted co-localization of spike S1 with sodium-potassium ATPase and F4/80, suggesting its presence on plasma membranes and within tissue macrophages (Figure S2I). In a time course experiment, we found the spike protein in the skull marrow, kidney, liver, and lung 3 days post-injection, remaining detectable in the kidney and liver 14 days post-injection (Figure S2J).
To explore the molecular consequences of SARS-CoV-2 infection and spike protein accumulation, we performed proteomics analysis of region-matched human tissues from COVID-19 patients and non-COVID-19 controls (Table S1).
Principal-component analysis (PCA) clearly separated skull marrow samples of COVID-19 patients from controls (Figure S3A). Gene set enrichment analysis indicated a strong association of differentially regulated proteins with the COVID-19 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (Figure 3A). Several proteins linked to the interleukin (IL)-18 signaling pathway were notably differentially expressed, including COL3A1, associated with lung fibrosis59 and ARF6, suggested to mediate viral entry.60 We also observed expression changes in proteins related to the MAPK and phosphatidylinositol 3-kinase (PI3K)-AKT signaling pathways and in proteins involved in neutrophil extracellular trap (NET) formation, such as HMGB1 and MPO (Figures 3B and S3B).
Figure 3 Proteomic changes in the skull marrow, meninges, and brain after SARS-CoV-2 infection in human and mouse samples
(A–F) Proteomic analysis of human samples. (A) KEGG pathway analysis showing the most dysregulated pathway in human COVID-19 skull marrow compared with non-COVID-19 controls. Dysregulated proteins related to NETs, IL-18 signaling, and PI3K-AKT signaling are listed in (B), n = 10. (C) Chord diagram showing the most enriched biological processes with their differentially expressed proteins (DEPs) in the COVID-19 meninges. n = 10. (D) Chord diagram showing the most enriched biological processes with their DEPs in the COVID-19 brain cortex. n = 8 COVID-19 patients, n = 11 non-COVID-19 controls. (E) Illustration of proteomics analysis of spike protein-positive and -negative brain regions from COVID-19 patients. (F) Volcano plot showing DEPs between spike protein-positive and -negative regions (p < 0.05, log2FC = 0.6). n = 3 COVID-19 patients. FC, fold change.
(G–N) Proteomic analysis of mouse samples. (G–I) Volcano plots of DEPs in the mouse skull marrow (G), meninges (H), and brain cortex (I) at 5 dpi (p < 0.05, log2FC = 1.5). n = 4 K18-hACE2 mice. (J) Heatmap showing the DEPs in the mouse brain cortex at 5 dpi. (K–M) Volcano plots of DEPs in the mouse skull marrow (K), meninges (L), and brain cortex (M) at 28 dpi (p < 0.05, log2FC = 1.5). n = 4 K18-hACE2 mice. (N) KEGG pathway analysis showing the most dysregulated pathways in the mouse brain cortex at 28 dpi compared with control Mock infection.
In the human meninges, PCA also effectively distinguished meninges samples between COVID-19 and control groups (Figure S3C). Notably, proteins involved in neutrophil degranulation were prominently differentially expressed (Figure 3C). Analyzing the skull and meninges samples together identified 1,460 upregulated and 26 downregulated proteins in COVID-19 patients compared with non-COVID-19 controls (Figures S3D and S3E), with proteins associated with the innate immune system and neutrophil degranulation notably elevated (Figure S3F).
Analysis of brain cortex samples revealed key affected pathways including complement activation, NET formation, coronavirus disease, and interferon (IFN)-alpha/beta signaling (Figure 3D). Comparing datasets from skull marrow, meninges, and brain samples revealed five proteins that were consistently differentially expressed in all three tissues: IGHV1-3, HLA-B, APCS, ISG15, and MX1 (Figure S3G).
To investigate the specific effects of spike protein in brain tissue beyond global inflammatory responses, we performed targeted proteomics on laser capture micro-dissected tissues61 by analyzing spike protein-positive versus spike protein-negative brain regions (Figure 3E), revealing dysregulated proteins linked to neurological diseases (Figure 3F). For instance, GFAP, a biomarker for BBB damage in COVID-19,62 was upregulated in spike protein-positive regions. Conversely, myelin basic protein (MBP), associated with multiple sclerosis,63 was downregulated in these regions. We also observed downregulation of GRIA1, a protein linked to attention deficits and sleep disorders—symptoms reported in post-COVID-19 syndrome.64,65,66
The dysregulation of proteins associated with neurological diseases after SARS-CoV-2 infection prompted our investigation into the shared proteomic alterations in the brain cortex between COVID-19 and Alzheimer’s disease (AD) (Figure S3H; Table S4). We identified 76 commonly dysregulated proteins in both COVID-19 and AD-affected brains (Figure S3I). We found key proteins with known roles in AD development: e.g., increased expression of GFAP67 and of PLXNC1, which is elevated in the microglia of human AD patients and mouse models.68 The common downregulated proteins included MAP6, a microtubule-associated protein critical for neuronal connectivity,69 NTM, which promotes neurite outgrowth,70 and several Rab proteins crucial for vesicle trafficking.71,72
To further investigate the impact of SARS-CoV-2 infections under more controlled conditions than in the human samples analyzed above, we determined the proteome of the skull marrow, meninges, and brain cortex of K18-hACE2 mice following SARS-CoV-2 (Wuhan-Hu-1 strain) infection.
At 5 days post-infection (dpi), 22 proteins were upregulated and 3 proteins downregulated in the skull marrow (Figure 3G). In the meninges, 27 proteins were upregulated and five downregulated (Figure 3H). Notably, 4 proteins—Serpina3, Ifit1, Itih3, and Itih4—were upregulated in both the skull marrow and meninges, and they have also been reported in the sera of COVID-19 patients.73,74 In the brain cortex, 12 proteins showed increased levels, with none decreasing at this early time point (Figures 3I and 3J). Importantly, the protein Itih4 was upregulated across all three regions, suggesting its involvement in inflammatory responses.75
At 28 days post-infection, 15 proteins were upregulated, and one was downregulated in the skull marrow (Figure 3K). 62 proteins demonstrated upregulation and three showed downregulation in the meninges (Figure 3L). In the brain cortex, 55 proteins were upregulated, and 225 proteins were downregulated (Figure 3M). Notably, the differentially regulated pathways were most strongly associated with neurodegeneration (Figure 3N). We also observed a persistent dysregulation of cytokine and other immune-related pathways (Figure 3N).
The temporal pattern of the proteome changes suggests a delayed impact of the viral infection on the brain proteome, as the brain exhibited fewer dysregulated proteins at 5 days post-infection than the skull/meninges, with a substantial increase observed at 28 dpi.
Given the persistent presence of spike protein in the skull, meninges, and brain cortex after SARS-CoV-2 infection, we investigated whether spike protein alone could trigger pathological effects.
We microinjected spike S1 protein into the skull marrow (Figures 4A and 4B)39 and found its rapid dissemination to the meninges and brain parenchyma within 30 min (Figure 4C). We assessed cell death and neuronal damage in brain cortex via the level of cleaved caspase-3 and amyloid precursor protein (APP),76,77 respectively. Notably, spike S1 induced a significant increase of cleaved caspase-3 at 28 days after injection, while HA only increased levels of cleaved caspase-3 at 3 days after injection (Figure 4D). Additionally, spike S1 induced a significant increase in APP expression in the brain at both time points, while HA injection did not elicit any changes (Figure 4E).
Figure 4 SARS-CoV-2 spike protein alone induces neuroinflammation
(A) Illustration of intra-skull microinjection in WT mice, accompanied by a 3D reconstruction showing the distribution of a red fluorescent dye within the skull post-injection.
(B) Representative images of mouse skull 30 min after injection of spike S1 (N501Y) and PBS control.
(C) Representative images of mouse brain cortex 30 min after injection.
(D) Representative images of c-caspase-3 antibody staining in mouse brain cortex after intra-skull injection of spike S1 (N501Y), HA protein, and PBS control and intensity-based quantification of caspase-3 activation. Each data point represents quantification from a distinct field of view. n = 3. One-way ANOVA with multiple comparisons.
(E) Representative images of APP antibody staining in mouse brain cortex after intra-skull injection and intensity-based quantification of APP. Each data point represents quantification from a distinct field of view. n = 3. One-way ANOVA with multiple comparisons.
(F) Schematic representation of WT mouse organotypic brain slice culture system.
(G) Representative images of the organotypic brain slices with/without pre-treatment of ACE2 antibody and treatment of spike S1 (N501Y).
(H) Quantification of CD68 and Iba1 fluorescent intensity. n = 4. Two-way ANOVA with multiple comparisons.
(I) Western blotting of the organotypic brain slice lysates to detect ERK and JNK phosphorylation.
(J) Quantification of phospho-ERK and phospho-JNK. n = 3. Two-way ANOVA with Bonferroni’s multiple comparisons. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns, nonsignificant.
To explore the mechanisms underlying spike-induced brain damage, we applied spike protein directly to organotypic brain slices (Figure 4F). We observed significantly increased levels of myeloid markers CD68 and Iba1 in the slices 3 days post-treatment with spike S1 protein (Figure S4A), indicating lysosomal activation and neuroinflammation. Control experiments using heat-denatured spike S1 protein showed minimal myeloid cell activation, with no significant differences in CD68 and Iba1 levels compared with vehicle treatment (Figure S4A). Furthermore, we found increased levels of IFN-gamma and CXCL10, but not IL-6, in the culture media after spike S1 treatment. No changes were observed with the denatured spike protein (Figure S4B).
Investigating the signaling cascades triggered by spike protein, we observed specific elevations in phosphorylated p44/42 MAPK (extracellular signal-regulated kinase [Erk] 1/2) and phosphorylated stress-activated protein kinase (SAPK)/JNK 1 h after spike protein, but not denatured spike protein, treatment, suggesting functional activation of the MAPK-JNK pathway and subsequent neuroinflammation induced by spike protein (Figures S4C and S4D). Pre-treatment with ACE2 antibody did not prevent the upregulation of CD68 but effectively reduced Iba1 intensity (Figures 4G and 4H) and inhibited the increase in p-p44/42 MAPK (Erk1/2) and p-SAPK/JNK (Figures 4I and 4J). These findings suggest that the neuroinflammatory effects of the spike protein are ACE2 dependent.
To further characterize (virus-independent) effects of the spike protein alone on the skull/meninges/brain axis, we performed proteomics analysis on day 3 post-intravenous injection of N501Y spike S1 protein, HA protein, and saline controls in WT mice.
The PCA plot showed skull marrow samples after spike S1 injection separated distinctly from the control group (Figure S5A). By contrast, the HA group partially overlapped with the control (Figure S5A). Compared with the saline control, we identified 808 differentially expressed proteins (DEPs) in the skull marrow after spike S1 injection (Figure S5B). In the skull marrow after HA injection, we only identified 16 DEPs compared with the control (Figure S5C). We found substantial differential expression of ribosomal proteins and of proteins involved in NETs formation, neutrophil degranulation, and PI3K-AKT pathways (Figure 5B). These pathways were also significantly dysregulated in COVID-19 patient skulls.
Figure 5 Circulating SARS-CoV-2 spike protein triggers proteomic changes in the mouse skull/meninges/brain and anxiety-like behavior
(A and B) (A) KEGG pathway analysis shows the most dysregulated pathways in mouse skull marrow after spike S1 (N501Y) injection, and the DEPs for selected pathways are listed in (B).
(C and D) (C) The most dysregulated pathways in the meninges, with the DEPs for selected pathways listed in (D).
(E and F) (E) The most dysregulated pathways in the brain cortex and the most enriched biological processes with their DEPs are shown in (F). n = 4 WT mice.
(G) Total distance traveled, time spent in center, and velocity of the mice in an open field during 20 min.
(H) Heatmap visualization representing the mouse position in the experimental arena. Red: more time, blue: less time.
(I–K) (I) Representative heatmap of the novel object recognition test. Quantification of the object exploration time at 3 and 28 dpi is shown in (J) and (K). Data are shown as mean ± SD. n = 15 WT mice. Two-way ANOVA with Bonferroni’s multiple comparisons, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, nonsignificant.
Similarly, the meninges samples after spike S1 injection distinctly separated from the control group in the PCA plot, whereas the HA group partially overlapped with the control (Figure S5D). We identified 426 DEPs after spike S1 injection (Figure S5E). Pathway analysis suggested the most abundant dysregulation in proteins involved in the formation of NETs. We also found proteins enriched in biological processes such as neutrophil degranulation, PI3K-AKT signaling pathway, and platelet activation, signaling, and aggregation (Figures 5C and 5D). We only identified 33 DEPs in the HA group compared with the control (Figure S5F).
In the brain cortex, the spike S1 group again showed a clear segregation from the HA and control group in the PCA plot (Figure S5G). 213 DEPs were identified in the brain cortex after spike S1 injection (Figure S5H). In addition to the altered level of ribosomal proteins associated with COVID-19, we identified proteins dysregulated in pathways of neurodegeneration (Figures 5E and 5F). GRIA1 and NDUFA3 were also dysregulated in the spike protein-positive COVID-19 patient brain regions (Figure 3F). After HA injection, only one protein was differentially expressed in the cortex (Figure S5I).
In blood plasma, both spike S1 and HA protein increased the inflammatory cytokines IFN-gamma and CXCL10. Plasma IL-6 levels were increased 3 days after spike S1 injection but remained unchanged after HA injection (Figure S5J).
To assess the functional consequences of spike protein presence, we evaluated mouse behavior 3 days post-injection. The mice spent less time and moved shorter distances in the center of an open field, suggesting spike protein-induced anxiety-like behavior. Walking velocity was not altered, indicating no motor deficits. The anxiety-like behavior was absent at 28 days post-injection (Figures 5G and 5H). In a novel object recognition test, no differences between the groups were observed (Figures 5I–5K).
Spike-induced long-term proteome changes at 28 dpi might exacerbate the effects of additional injuries to the brain. To test this, we used two distinct brain injury models 35 days after intravenous injection of spike protein and a saline control: middle cerebral artery occlusion (MCAo)-induced stroke and TBI induced by controlled cortical impact (CCI) (Figure S6A).
For the MCAo model, Neuroscore78 assessments revealed increased deficits in whisker and forelimb symmetry in mice with spike protein injection (Figure 6A). The volume of the brain infarct region showed no significant change between mice with and without spike injection at 24 h after MCAo (Figure 6B). Proteomic analysis of the peri-infarct brain region (Figures 6C and S6B) unveiled significant increases in proteins such as CD200, a key regulator of immune response in the brain79; Efnb2, involved in axon guidance and synaptic plasticity80; and ASTN2, associated with the trafficking and degradation of synaptic proteins81 in spike S1-injected animals. Conversely, Gphn, a component of inhibitory synapses,82 was decreased in the brains of mice after spike injection and MCAo.
Figure 6 SARS-CoV-2 spike protein exacerbates neurological damage
(A) Individual Neuroscore assays of the mouse 24 h after MCAo.
(B) Nissl staining of brain sections 24 h after MCAo with quantification of infarct volume.
(C) DEPs in the peri-infarct region at 24 h after MCAo.
(D–G) (D) MRI scan of the mouse brain after traumatic brain injury (TBI). Lesion volume (E), midline shift (F), and the ratio of ipsilateral to contralateral hemisphere volume (G).
(H) Quantification of brain tissue loss after the acute phase of injury.
(I) Quantification of the lateral ventricle volume.
(J) Illustration of the brain region for proteomic and histology analysis at 48 days after TBI.
(K) DEPs in mouse brain 48 days after TBI.
(L) Representative images of CD68 and GFAP staining in the corpus callosum.
(M) Representative images of c-caspase-3 and Nissl staining in the corpus callosum. Data are shown as mean ± SD. n = 5 WT mice, unpaired two-tailed t test, ∗p < 0.05, ns, nonsignificant.
In the TBI model, sequential MRI scans depicted substantial cortical lesions and brain hemisphere swelling during the acute-phase (4–72 h) in T2 weighted scans, which gradually subsided in the subacute-phase (days 7–14) (Figure 6D). Although there was no significant difference in total lesion volume or midline shift between mice with and without spike protein injection (Figures 6E and 6F), mice with spike protein injection exhibited significantly decreased ipsilateral hemisphere volume (Figure 6G) at 42 days post-TBI and significantly increased brain tissue loss at 14 and 42 days post-TBI (Figure 6H), suggesting spike protein exposure led to more brain atrophy in the chronic-phase of TBI. Chronic-phase observations at 42 days after TBI also revealed significantly increased lateral ventricle volume in mice with spike protein injection (Figure 6I), suggesting potential obstruction of CSF flow and a propensity for more hydrocephalus compared with saline controls.
Upon sacrifice at 48 days after TBI, brain proteome analysis (Figures 6J and S6C) revealed heightened expression of proteins relevant to inflammatory pathways, including inhibitor of nuclear factor kappa B (IκB) phosphorylation and the tumor necrosis factor (TNF) signaling pathway (Figure S6D) in spike protein-injected mice. Notably, Pdk2, known for contributing to oxidative glucose metabolism in TBI,83 exhibited increased expression in brains with spike protein injection after TBI (Figure 6K). Immunofluorescence imaging showed TBI-induced diffuse axonal injury in the white matter was accompanied by elevated levels of CD68 and c-caspase-3 in the corpus callosum of mice with spike protein injection, indicating increased neuroinflammation and ongoing cell death in the mouse brain after TBI (Figures 6L and 6M).
We further investigated the protective efficacy of vaccination against both virus infection and the extended distribution of the spike protein in mice. Given that the Omicron XBB.1.5 variant of SARS-CoV-2, which became predominant in the world in 2023,84 can directly infect WT mice, providing a more accurate representation of infection and viral distribution compared with the ancestral Wuhan strain in transgenic K18-hACE2 mice, we conducted this experiment using WT mice and the Omicron XBB.1.5 variant. We administered two doses of the BioNTech/Pfizer Comirnaty vaccine intramuscularly and aerosol-infected mice with the Omicron XBB.1.5 10 days after completing the full vaccination regimen. No body weight loss was observed in vaccinated and non-vaccinated mice after XBB.1.5 infection (Figure S7A). At 5 days post-infection, the mice were sacrificed, and we conducted whole-body clearing and imaging to visualize the distribution of the spike protein (Figure 7A).
Figure 7 The accumulation of spike protein after SARS-CoV-2 infection is reduced with Comirnaty vaccination
(A) 3D reconstruction and optical section of spike protein distribution in WT mouse body with/without vaccination and 5 days after Omicron XBB.1.5 infection.
(B) Zoomed-in images showing spike protein in different organs.
(C) Representative confocal images of spike and nucleocapsid protein in the mouse brain.
(D) Quantification of nucleocapsid and spike protein in the mouse brain. Data are shown as mean ± SD, n = 3, unpaired two-tailed t test, ∗p < 0.05, ∗∗p < 0.01.
(E) Serum IL-6 level in WT mice with/without vaccination and Omicron XBB.1.5 infection. Pooled data of 2 independent experiments (n = 4 non-vaccinated, n = 8 vaccinated). One-way ANOVA with Bonferroni’s multiple comparisons.
Compared with the unvaccinated animals, we observed lower spike protein levels in the lungs, liver, kidneys, hearts, skull marrow, and brains of vaccinated mice (Figures 7B and S7B). Both spike and nucleocapsid proteins were also significantly decreased in the brain cortex of vaccinated mice (Figures 7C and 7D), though some vaccinated mice still show lingering spike protein, suggesting a balance between viral replication and clearance. The detection of viral proteins in the brain following mild Omicron infection suggests it may contribute to long-term memory deficits, complementing other mechanisms such as heightened peripheral inflammation observed with more severe variants.85
Lung tissue lysates confirmed that all unvaccinated mice were positive for the virus, while vaccinated animals cleared the infection, validating the infection and vaccination efficiency in our experiments (Figure S7C). Serum analysis for spike immunoglobulin G (IgG) levels further confirmed effective vaccination, XBB.1.5 infection alone did not induce detectable spike IgG in mouse serum at 5 dpi (Figure S7C). We also assessed serum levels of IL-6 and TNF-α, finding significantly reduced IL-6 levels in vaccinated mice compared with non-vaccinated mice after infection (Figures 7E and S7D). These results suggest that vaccination effectively reduces both inflammation and viral protein accumulation in mice following infection, and this aligns with large cohort studies showing that vaccination reduces the risk of long COVID and its associated symptoms.86,87
The long-term complications of COVID-19 remain a major public health concern, and the long-lasting brain fog and significant brain tissue loss make it imperative to explore the mechanisms of SARS-CoV-2-induced brain damage.7,8,88 Increased plasma cytokine levels, blood-brain barrier disruption, and reactivation of latent herpesviruses have been correlated with cognitive symptoms associated with long COVID.8,85,89,90,91 The persistence of spike protein is also reported in patient plasma30,92 and immune cells,29 suggesting that the prolonged symptoms post-COVID-19 could stem from the enduring presence of viral proteins, alongside sustained systemic inflammation. Here, using tissue clearing and imaging, we detected spike protein in skull, meninges, and brain samples from acute COVID-19 patients and reported the persistence in post-mortem brain samples that were PCR negative for the virus, which may explain some aspects of COVID-19 symptoms.93 Visualizing the whole-body distribution of SARS-CoV-2-GFP virus infection in K18-hACE2 mice and of a mouse ACE2-binding spike S1 protein (with N501Y mutation) in WT mice, we found the expected target organs, such as the lung, kidney, and liver,51,94,95 and observed the accumulation of spike protein in the skull marrow and SMCs, similar to our findings in the humans.
We found that the spike protein alone induced a broad spectrum of proteome changes in the mouse skull marrow, meninges, and brain that are similar to those observed in COVID-19 patients and that the isolated effects of the spike protein on the nervous system caused anxiety-like behavior without memory deficits. While previous studies reported anxiety and cognitive impairment induced by direct administration of spike protein into the hippocampus or ventricles,27,96 the differences between our and previous studies are likely caused by the higher local concentration of spike protein in the hippocampus in those studies. The worsened outcomes after MCAo and TBI indicated that the spike protein may render the brain more vulnerable to subsequent insults. No significant difference was observed in the TBI model during the acute phase. However, a more pronounced injury attributable to spike treatment was detected in the chronic phase. Our MCAo assessment focused solely on the acute phase, and additional long-term spike protein-induced changes may not have been observed. While these findings do not support the direct induction of clinical neurological symptoms by the spike protein, they suggest the long-term consequences of spike protein-induced inflammation and dysfunctional signaling in the brain. Further investigation is needed to elucidate the impact of spike protein on the brain and its association with other neurological diseases.
Our data also suggest a mechanism for the virus’s entry into the central nervous system. In both mouse and COVID-19 human tissues, we found spike protein in the SMCs, which the virus or virus components could use to travel from the skull marrow to the meninges and the brain parenchyma.34,39,40,41 Brain entry of virus-shed spike protein has previously been found in some COVID-19 cases and has been linked to a compromised blood-brain barrier32,97 or trafficking along the olfactory or vagus nerve.9 More data will be needed to establish the relative contribution of the different routes of brain invasion by SARS-CoV-2 and its spike protein, which might differ between different parts of the brain.
Systemic hyperinflammation has previously been observed in multiple organs during autopsies of COVID-19 cases.98 Our molecular analysis suggests activation of immune response in the skull-meninges-brain axis, potentially via recruiting and increasing the activity of neutrophils similar to what has been reported in the respiratory tract.99,100 In the skull marrow and meninges, the upregulation of proteins involved in neutrophil degranulation may be linked with a process known as NETosis to contain the infection. NETs could further propagate the inflammation,101 potentially inducing tissue damage, including endothelium damage,102 leading to pathologies such as thrombosis and alterations in the coagulation process,103 as alterations in such processes are found in lung tissue and plasma of COVID-19 patients.104,105 Targeting NETs has reduced multiple organ injury in SARS-CoV-2 infected mouse model.106 In brain cortex tissue, our data highlighted dysregulated proteins that were also identified in previous transcriptomic studies of COVID-19 affecting the brain and choroid plexus,14 including IFITM3, STAT3, C7, NQO1, ZFP36, SDC4, C3, and OSMR. Most of these proteins are involved in immune response, inflammation, and cellular stress responses. Furthermore, VIM, VCL, MYH2, COL6A3, and MYH11, identified as dysregulated in the COVID-19 brain cortex, have also been documented in other organs, including the lung, kidney, liver, and intestine,107 and these proteins play pivotal roles in cytoskeleton dynamics, cell adhesion, migration, and the preservation of tissue integrity.
Spike-induced alterations in the skull-meninges-brain axis present diagnostic and therapeutic opportunities as both skull and meninges are easier to access than brain parenchyma. Panels of such proteins tested in plasma or CSF samples of COVID-19 patients might provide an early prognosis of brain-related complications. Future efforts should be committed to characterizing these proteins toward their use as biomarkers and therapeutic targets for neurological dysfunction in COVID-19.
Our study has several limitations. First, the assessment of human tissues is constrained by its sample size, and larger independent cohorts are needed to explore comorbidities. Our human skull and brain tissue samples focus mainly on the frontal-parietal region, and the samples for proteomic analysis of the skull and brain were not consistently derived from the same patients (Table S1). Second, although elevated neurodegenerative biomarkers were detected in CSF of long COVID patients, we could not assess spike protein presence in their skull-meninges-brain tissues due to a lack of post-mortem tissue samples, preventing us from correlating spike protein persistence with clinical neurological symptoms. Additionally, we lack information on the SARS-CoV-2 variants in the studied patients. Given the different transmission and pathological profiles of each variant, further research is essential to assess the impact of mutations on the persistence of spike protein. Moreover, the findings from the mouse models do not fully recapitulate the viral tropism of SARS-CoV-2 in humans. Particularly, the ectopic expression of hACE2 in K18-hACE2 mice might lead to changes in tissue tropism, including increased brain infection, that might not accurately reflect human pathology. Lastly, while we demonstrated spike protein persistence and immune activation in the skull-meninges-brain axis, future clinical studies are needed to investigate the clinical effects of reducing spike protein levels in long COVID patients, complementing the preventive effects of vaccination.
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ali Ertürk ([email protected]).
This study did not generate new, unique reagents.
We thank Alireza Ghasemi for developing the Python script to stitch sequences of images. We thank Christoph Krisp (Bruker Daltonics) for helpful discussions on mass spectrometer data acquisition. We thank Mrs. Brigitte Nuscher for her assistance with the Simoa assay. Illustrations were created with BioRender.com. Z.R. and H.M. thank the China Scholarship Council (CSC) for the financial support (no. 201806310110 and no. 201806780034). We acknowledge all families supporting our research after losing their beloved ones during the pandemic. This work was supported by the European Research Council Consolidator grant (CALVARIA, grant no. GA 865323 to A.E.), the NOMIS Human Heart Atlas Project grant (Nomis Foundation), the Vascular Dementia Research Foundation, the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy within the framework of the Munich Cluster for Systems Neurology (EXC 2145 SyNergy, ID 390857198), and the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung, BMBF) within the NATON collaboration (01KX2121) and DFG (SFB 1052, project A9; TR 296 project 03; TRR353/B02 to G.E.). This work was also supported by the State of Bavaria and the European Union via a grant for regional infrastructure development (EFRE-REACT to U.P. and G.E.) by the Helmholtz Association’s Initiative and Networking Fund (KA1-Co-06, “CORAERO” to G.E.) and a GoBio project (16LW0286K to G.E. and U.P.). G.H., K.-W.S., M.N., and F. Hopfner are supported through the COFONI-2FF4/COVID-19-Research Network of the State of Lower Saxony (COFONI), Ministry of Science and Culture of Lower Saxony, Germany.
A.E. conceived and led all aspects of the project. Z.R. and H.M. designed and carried out most of the experiments. G.E. and D.M. performed mouse infection experiments. S.K., M.A., S.U., N.K., Ö.S.C., and M.M. performed mass spectrometry-based proteomic experiments. Z.R., H.M., G.E., and S.K. performed data analysis and visualization. V.G.P., J. Schädler, T.B.H., and B.O. organized human sample collection. J.C. dissected the human skull with the dura mater sample and collected clinical information. S.H. performed TBI experiments and MRI analysis. J. Su and U.P. conducted Comirnaty vaccination. D.P. performed organotypic mouse brain slice culture. I.S. performed western blotting. C.D., H.S., H.F., K.S., C.B., G.B., and I.B. provided human brain and skull samples. V.R. and J.H. provided human brain samples of AD, COVID-19, and control patients from the Neurobiobank Munich. K.-W.S., M.N., and F. Hopfner provided and analyzed CSF samples. D.J. performed MCAo experiments. J.M.W. performed quantitative reverse-transcription PCR (RT-qPCR) analysis. F.C. assisted in sample homogenization. Z.I.K. helped with human skull sample collection. I.H. and S.Z. participated in prototyping experiments. A.E., F. Hellal, and H.S.B. supervised the project. Z.R., H.M., S.K., M.E., and A.E. wrote the manuscript. All authors reviewed and approved the final manuscript.
The authors declare no competing interests.
REAGENT or RESOURCESOURCEIDENTIFIERAntibodiesSARS-CoV-2 (COVID-19) Spike antibody (dilution for immunofluorescent staining 1:1000)GeneTexCat# GTX135356; RRID: AB_2887482SARS-CoV / SARS-CoV-2 (COVID-19) spike antibody [1A9] (dilution for immunofluorescent staining 1:1000)GeneTexCat# GTX632604; RRID: AB_2864418FluoTag®-Q anti-S1 SARS-CoV-2 AZDye568 (dilution for immunofluorescent staining 1:1000)NanoTag BiotechnologiesCat# N3501-AF568-L; RRID: AB_3076086GFP-Booster Alexa Fluor 647 (dilution for immunofluorescent staining 1:1000)ChromoTekCat# gb2AF647; RRID: AB_2827575SARS Coronavirus Nucleocapsid Polyclonal Antibody (dilution for immunofluorescent staining 1:1000)Thermo FisherCat# PA1-41098; RRID: AB_1087200ACE2 Polyclonal Antibody (dilution for immunofluorescent staining 1:1000)Thermo FisherCat# PA5-20039; RRID: AB_11154831Anti Iba1, Rabbit (for Immunocytochemistry) (dilution for immunofluorescent staining 1:1000)WakoCat# 019-19741; RRID: AB_839504Influenza A H1N1 haemagglutinin antibody [C102 (IV.C102)] (dilution for immunofluorescent staining 1:1000)AbcamCat# ab128412; RRID: AB_11142609Collagen IV Antibody, anti-human, PE, REAdye_lease™ (dilution for immunofluorescent staining 1:500)Miltenyi BiotecCat# 130-122-866; RRID: AB_2857566CD31 antibody (dilution for immunofluorescent staining 1:1000)AbcamCat# ab32457; RRID: AB_726369Rat Anti-CD31 Monoclonal Antibody, Unconjugated, Clone MEC 7.46 (dilution for immunofluorescent staining 1:1000)AbcamCat# ab7388; RRID: AB_305905NeuN Polyclonal Antibody (dilution for immunofluorescent staining 1:500)Thermo FisherCat# PA5-78499; RRID: AB_2736206Cleaved Caspase-3 (Asp175) (5A1E) Rabbit mAb (dilution for immunofluorescent staining 1:200)Cell Signaling TechnologyCat# 9664; RRID: AB_2070042beta Amyloid Polyclonal Antibody (CT695) (dilution for immunofluorescent staining 1:500)InvitrogenCat# 51-2700; RRID: AB_2533902F4/80 antibody [CI:A3-1] (dilution for immunofluorescent staining 1:1000)AbcamCat# ab6640; RRID: AB_1140040Rabbit Anti-Sodium Potassium ATPase Monoclonal Antibody, Unconjugated, Clone EP1845Y (dilution for immunofluorescent staining 1:500)AbcamCat# ab76020; RRID: AB_1310695Neuronal Class III beta-Tubulin (TUJ1) Monoclonal Antibody, Purified (dilution for immunofluorescent staining 1:500)CovanceCat# MMS-435P; RRID: AB_2313773RAT ANTI MOUSE CD68 (dilution for immunofluorescent staining 1:1000)Bio-RadCat# MCA1957GA; RRID:AB_324217Anti-IBA1 (dilution for immunofluorescent staining 1:500)Synaptic SystemsCat# 234 308; RRID:AB_2924932Recombinant Anti-PDGFR beta antibody [Y92] – C-terminal (dilution for immunofluorescent staining 1:500)AbcamCat# ab32570; RRID: AB_777165Mouse Anti-Aquaporin 4 Monoclonal Antibody, Unconjugated, Clone Apr 18 (dilution for immunofluorescent staining 1:500)AbcamCat# ab9512; RRID: AB_307299Anti-Tyrosine Hydroxylase Antibody (dilution for immunofluorescent staining 1:500)Merck MilliporeCat# AB152; RRID: AB_390204Goat Anti-Rabbit IgG H&L (HRP) (dilution for immunohistochemistry staining 1:1000)AbcamCat# ab6721; RRID: AB_955447Goat Anti-Rabbit IgG (H+L) Highly Cross-adsorbed Antibody, Alexa Fluor 647 Conjugated (dilution for immunofluorescent staining 1:1000)Thermo FisherCat# A-21245; RRID: AB_141775Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 568 (dilution for immunofluorescent staining 1:1000)Thermo FisherCat# A-11036; RRID: AB_10563566Goat Anti-Mouse IgG (H+L) Highly Cross-adsorbed Antibody, Alexa Fluor 568 Conjugated (dilution for immunofluorescent staining 1:1000)Thermo FisherCat# A-11031; RRID: AB_144696Goat anti-Mouse IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 647 (dilution for immunofluorescent staining 1:1000)Thermo FisherCat# A-21235; RRID: AB_2535804Goat anti-Rat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 568 (dilution for immunofluorescent staining 1:1000)Thermo FisherCat# A-11077; RRID: AB_2534121SAPK/JNK Antibody (dilution for immunoblotting 1:1,000)Cell Signaling TechnologyCat#9252; RRID: AB_2250373Phospho-SAPK/JNK (Thr183/Tyr185) Antibody (dilution for immunoblotting 1:1000)Cell Signaling TechnologyCat#9251; RRID: AB_331659Anti-p38 MAPK Antibody, Unconjugated (dilution for immunoblotting 1:1000)Cell Signaling TechnologyCat#9212; RRID: AB_330713Phospho-p38 MAPK (Thr180/Tyr182) Antibody (dilution for immunoblotting 1:1000)Cell Signaling TechnologyCat#9211; RRID: AB_331641p44/42 MAP kinase (phosphorylated Erk1/2) (dilution for immunoblotting 1:1000)Cell Signaling TechnologyCat#9101; RRID: AB_331646p44/42 MAPK (Erk1/2) Antibody (dilution for immunoblotting 1:1000)Cell Signaling TechnologyCat#9102; RRID: AB_330744Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP (dilution for immunoblotting 1:5000)InvitrogenCat#31460; RRID: AB_228341Bacterial and virus strainsSARS-CoV-2-GFPStukalov et al.48N/ASARS-CoV-2 Wuhan-Hu-1Stukalov et al.48N/ASARS-CoV-2 Omicron (B.1.1.529, sub-lineage XBB.1.5)Stukalov et al.48N/ABiological samplesHuman skull and meninges samples from COVID-19 patients and controlsThis study; University Medical Center Hamburg-EppendorfN/AHuman skull samples from donors died of non-COVID-19 causes during 2021-2022This study; Institute of Pathology, Technical University of MunichN/AHuman brain and meninges samples from COVID-19 patients and controlsThis study; Anatomical Institute of the University of LeipzigN/AHuman cerebrospinal fluid from SARS-CoV-2-infected or uninfected donorsThis study; Medical School HannoverN/AHuman frontal cortex brain blocks from COVID-19, Alzheimer’s Disease, and control donorsThis study; Neurobiobank Munich (NBM)https://www.en.neuropathologie.med.uni-muenchen.de/neurobiobank/neurobiobank/index.htmlChemicals, peptides, and recombinant proteinsSARS-CoV-2 (COVID-19) S1 protein (N501Y), His TagAcrobiosystemsCat# S1N-C52HgInvitrogen Influenza A H1N1 HA (A/California/04/2009) His-tag ProteinThermo Fisher ScientificCat# A42579Coronavirus (COVID-19 Spike Protein, Full Length) AntigenEastCoast BioCat# LA636PBSRatiopharmCat# N68542.034% paraformaldehyde (PFA)MorphistoCat# 11762.05000Propidium iodide (PI)Sigma-AldrichCat# P4864Triton X-100PanReac ApplichemCat# A4975,1000Goat serumGIBCOCat# 16210072TRITC-DextranSigma-AldrichCat# 52194DMSORothCat# A994.2Tetrahydrofuran (THF)RothCat# CP82.1Dichloromethane (DCM)Sigma-AldrichCat# 270997Benzyl benzoateSigma-AldrichCat# W213802Benzyl alcoholSigma-AldrichCat# 24122CHAPSCarl RothCat# 1479.4N-MethyldiethanolamineSigma-AldrichCat# 471828Goat serumLifeCat# 16210064Critical commercial assaysAlexa Fluor® 647 Conjugation KitAbcamCat# ab269823Allplex™ 2019-nCoV AssaySeegeneCat# RP10243XImmPRESS Duet Double Staining Polymer KitVectorlabsCat# MP-7714Neurology 4-Plex A assay kitQuanterixCat# 102153Deposited dataMass spectrometry raw data, libraries and outputsThis paperhttps://www.ebi.ac.uk/pride/; ID# PXD050384Experimental models: Organisms/strainsMouse: C57BL/6JJackson LaboratoryStrain #:000664; RRID: IMSR_JAX:000664Mouse: K18-hACE2 (B6.Cg-Tg(K18-ACE2)2Prlmn/J)Jackson LaboratoryStrain #:034860; RRID: IMSR_JAX:034860Software and algorithmsImageJ – FijiSchneider et al.108https://imagej.nih.gov/ij/Prism 8.0GraphPadhttps://www.graphpad.com/scientific-software/prism/Imaris 9.6Oxford Instrumentshttps://imaris.oxinst.com/MaxQuant (1.6.7.0)Tyanova et al.109https://maxquant.org/Perseus (1.6.7.0)Tyanova et al.109https://maxquant.org/perseus/
Mix-gender 2-month-old wildtype C57BL/6J mice and K18-hACE2 transgenic mice (B6.Cg-Tg(K18-ACE2)2Prlmn/J) were purchased from Charles River Laboratories (MA, USA) and Jackson Laboratory (ME, USA), respectively, and bred in pathogen-free animal facilities following institutional guidelines. Animals were housed on a 12/12 hr light/dark cycle and had arbitrary access to food and water. Temperature was maintained at 18-23 °C and humidity at 40-60%. Animal experiments were conducted in strict accordance with the German regulations of the Society for Laboratory Animal Science (GV-SOLAS), the European Health Law of the Federation of Laboratory Animal Science Associations (FELASA), and the European Directive 2010/63/EU for animal experiments. Experiments were approved by the District Government of Upper Bavaria (Regierung von Oberbayern, Munich, Germany) (permission numbers: ROB-55.2-2532.Vet_02-21-169 and ROB-55.2-2532.Vet_02-20-193). All data are reported according to the criteria of ARRIVE. The sample size was chosen based on previous experience with similar models.
PFA-fixed human brains and human skull blocks were obtained from donors and autopsies in case of a diagnosis of COVID-19 during a lifetime or positive SARS-CoV-2 qPCR test post-mortem and under the European Control for Infectious Diseases. All donors or next-of-kin gave their informed and written consent to examine the cadavers for research and educational purposes. Human skull samples were collected during autopsies at the Institute of Legal Medicine of the University Medical Center Hamburg-Eppendorf, Germany, and the Institute of Pathology of the Technical University of Munich, Germany. Human brain tissue was obtained from the Anatomical Institute of the University of Leipzig, Germany. The post-mortem interval was on average 5 d. Institutional approval was obtained following the 1994 Saxon Death and Burial Act and of the independent ethics committee of the Hamburg Chamber of Physicians (protocol 2020-10353-BO-ff). Human CSF samples were obtained from the Medical School Hannover, Germany. Human brain samples of AD, COVID-19, and control patients were obtained from the Neurobiobank Munich (NBM) (www.en.neuropathologie.med.uni-muenchen.de/neurobiobank/neurobiobank/index.html). Patient information can be found in the supplementary material (Tables S1–S4).
SARS-CoV-2 Omicron (B.1.1.529, sub-lineage XBB.1.5) and SARS-CoV-2-GFP48 were produced, and virus stock titers determined as described previously.48 For SARS-CoV-2-GFP studies, K18-hACE2 transgenic mice (B6.Cg-Tg(K18-ACE2)2Prlmn/J) were aerosol-inoculated with 1.2×106 TCID50 SARS-CoV-2-GFP in a total volume of 6 ml. For SARS-CoV-2 Omicron infection studies, C57BL/6J mice were aerosol-inoculated with 3×106 TCID50 Omicron XBB.1.5 in a total volume of 6 ml. For immunization before SARS-CoV-2 Omicron infection, C57BL/6J mice were kept in pathogen-free animal facilities following institutional guidelines and were injected intramuscularly with 2.4 μg of BNT162b2 mRNA vaccine (BioNTech/Pfizer) vaccine at weeks 0 and 4. Subsequently, mice were transferred to the BSL-3 facility for virus challenge. SARS-CoV-2 infected mice were monitored for clinical signs of disease and weighed daily. Mice were euthanized at the indicated time points by CO2 asphyxiation or isoflurane overdosing.
Spike S1 protein (N501Y), WT spike protein, and hemagglutinin (HA) were labeled with the fluorescent dye Alexa Fluor 647 according to the manufacturer’s protocol (Alexa Fluor 647 conjugation kit lightning link, Abcam, ab269823). Briefly, the candidate protein (20 μg) supplied by the manufacturer (SARS-CoV-2 (COVID-19) S1 protein (N501Y), His Tag (S1N-C52Hg, Acrobiosystems), HA Recombinant Influenza A Virus Protein, subtype H1N1 (A/California/04/2009) (A42579, Life Technologies), Coronavirus (COVID-19 Spike Protein; Full Length) Antigen, Recombinant >90% (LA636, EastCoast Bio)) were dissolved in 0.01 M PBS (18 μl, pH 7.4). The modifying reagent (1 μl) was added to the protein solution. The solution was then mixed gently and transferred to a lyophilized material. After 15 minutes of incubation at room temperature, the reaction was terminated by adding the quencher reagent (1 μl). The fluorophore-labeled protein had a concentration of 1 mg/ml.
Anesthesia was induced in the animals with 4% isoflurane in an N2O/O2 mixture (70%/30%) and then maintained with 1.5% isoflurane in the same mixture for the entire injection. A 0.1 ml PBS solution containing 10 μg Alexa-647 conjugated protein was injected into the mouse tail vein using a 1 ml syringe with a 28-gauge needle. Microinjection into the skull marrow was performed in mice under 1.5% isoflurane anesthesia using a 5 μl syringe (#87943, Hamilton) equipped with a 34-gauge blunt needle. The skin above the skull was opened at the midline to expose the anterior marrow sites (near the bregma). A drill bit was used as a pilot drill to remove the cortical portion of the skull and expose the spongy bone marrow. A microinjection was performed for each marrow site pre-drilled with a 30-gauge needle (#305106, BD) to prevent perforation of the inner skull wall. In this study, the red tracker (C34565, Molecular Probes) was diluted to 10 μM according to the manufacturer’s protocol. The Alexa Fluor 647 conjugated protein solution (1 mg/ml) was prepared, and a total volume of 2.5 μl solution was slowly injected per injection site through the pre-drilled sites (20-30 s per injection), resulting in a total of 10 μl of conjugated spike protein injected per skull. The injection process was closely monitored under a microscope. Animals in which the procedure was unsuccessful were removed from the study. After the injection, the skin was sutured with 6-0 silk. The mice were euthanized at given time points and the heart was then perfused with PBS and fixed with 4% PFA for further analysis.
Mice were deeply anesthetized by intraperitoneal injection with a combination of midazolam, medetomidine, and fentanyl (1 ml/100 g of body mass for mice) until the mice showed no response with a pedal reflex. Then, the mice were perfused intracardially with heparinized 0.01 M PBS (10 U/ml of Heparin, Ratiopharm, N68542.03; 100-125 mmHg pressure using a Leica Perfusion One system) for 5 minutes to remove the blood, followed by a total volume of 20 ml of dextran (5 mg/ml, MW 500000, Sigma, 52194) for vascular labeling. Mice were then perfused with 50 ml of 4% paraformaldehyde (PFA) in 0.01 M PBS (pH 7.4; Morphisto, 11762.01000) for fixation. Then, bodies were post-fixed in 4% PFA for 1 day or 5 days after infection respectively at 4°C and washed three times with 0.01 M PBS for 10 minutes at room temperature. The mice were then tissue-cleared using the 3DISCO method.
We performed tissue clearing based on the 3DISCO and vDISCO protocol for whole mice as previously described.33,34,35 For the tissue clearing of mice after intravenous injection, mouse bodies were placed in a 300 ml glass chamber and incubated in 200 ml of the following gradient of THF (Tetrahydrofuran, Roth, CP82.1) in distilled water in a fume hood with gentle shaking: 50% ×1, 70% ×1, 80% ×1, 100% ×2, 12 h for each step, followed by 3 h in dichloromethane (DCM, Sigma, 270997) and finally in BABB solution (benzyl alcohol + benzyl benzoate 1:2, v/v, Sigma, 24122 and W213802) until optical transparency. For the tissue clearing of mice after virus infection, the mouse bodies were placed inside a 300 ml glass chamber (Omnilab, 5163279), to be filled with the appropriate solution regarding the protocol to cover the entire animal body (250-300 ml). A transcardial-circulator system was established in order to allow peristaltic pumping of the solutions (ISMATEC, REGLO Digital MS-4/8 ISM 834; reference tubing, SC0266), with the pressure being set at 180-230 mmHg (50-60 rpm). The tubing was set to allow pumping of the solutions through the heart (attached to a perfusion needle (Leica, 39471024)) into the vasculature with the same entry point used for PBS and PFA perfusion steps described above. The other end of the tube was immersed into the chamber with a loose end to allow suction of the solution into the body. The samples were initially perfused with a decolorization solution (25% of CUBIC reagent 190, which is composed of 25 wt% urea (Carl Roth, 3941.3), 25 wt% N,N,N’,N’-tetrakis (2-hydroxypropyl)ethylenediamine (Sigma, 122262) and 15 wt% Triton X-100 (AppliChem, A4975,1000) in 0.01 M PBS) for 2 days, refreshing the solutions every 12h. Samples were washed with PBS for 3x2h. Then, decalcification solution (10 wt/vol% EDTA in 0.01 M PBS, pH 8-9, Carl Roth, 1702922685) was perfused for 2 days followed by half a day with permeabilization solution composed of 0.5% Triton X-100, 1.5% goat serum (GIBCO, 16210072), 0.5 mM of Methyl-beta-cyclodextrin (Sigma, 332615), 0.2% trans-1-Acetyl-4-hydroxy-L-proline (Sigma, 441562), 0.05% sodium azide (Sigma, 71290) in 0.01 M PBS. Then, the samples were perfused for 6 days after adding 35 μl of nanobooster, which was added directly into the refreshed permeabilization solution. Next, the body was placed into a 50 ml tube (Falcon, 352070), with the same permeabilization and labeling solution, and an extra 5 μl of nanobooster was added. The tube was then put on a shaker at RT for 2 additional days for labeling. GFP-Booster Alexa Fluor 647 (Chromotek, gb2AF647) and AF568 anti-S1 SARS-CoV-2 (NanoTag, N3501-AF568-L) were used to boost the signal from the GFP and spike protein respectively in the study. Then, the animals were placed back into the initial perfusion setup, where the washing solution was perfused for 2x12h, composed of 1.5% goat serum, 0.5% Triton X-100, and 0.05% sodium azide in 0.01 M PBS. 0.01 M PBS was used to wash the sample 3x2h. 3DISCO protocol was applied for whole-body clearing. The animals were freed from the perfusion system but kept in glass chambers and placed on top of shakers (IKA, 2D digital) at room temperature inside a fume hood. Glass chambers were sealed with parafilm and covered with aluminum foil along with the 3DISCO application. For dehydration, sequential immersion of tetrahydrofuran (THF) (Sigma, 186562) (50 Vol% THF, 70 Vol% THF, 80 Vol% THF, 100 Vol% THF, and again 100 Vol% THF) was applied every 12 hours. Then, three hours of DCM (Sigma, 270997) immersion for delipidation was followed by indefinite immersion in BABB solution for refractive index matching.
Deep tissue labeling was performed according to the protocol known in our laboratory as the SHANEL protocol.36 Cut 1 cm thick slices of the desired brain tissue or human skull before SHANEL pretreatment. Then, treat the human brain slices with CHAPS /NMDEA solution (10% CHAPS and 25% NMDEA in diH2O) twice at room temperature for 12 hours each time. Wash three times with PBS for 20 minutes each time. The tissue was dehydrated by stepwise addition of ethanol (50% ×1, 70% ×1, 100% ×1, 4 h for each step). Change to DCM/MeOH (2:1, v/v) solution overnight. Tissues were rehydrated stepwise in gradient ethanol (100% ×1, 70% ×1, 50%×1) and diH2O, 4 h for each step), treated overnight with 0.5 M acetic acid solution and washed twice for 20 min in diH2O, treated with guanidine solution for 6 h, and washed 2 more times in diH2O. Then block the tissue with blocking buffer (0.2% Triton X-100, 10% DMSO (Roth, A994.2), 10% goat serum in 0.01 M PBS) at 37 °C overnight and then incubate it with propidium iodide (1:1000, Sigma, P4864) or primary antibodies in antibody incubation buffer (3% goat serum, 3% DMSO, 0.2% Tween-20, 10 mg L-1 heparin in 0.01 M PBS) at 37 °C for 5 days. After incubation with the primary antibodies, the tissue was washed twice in PBS for 1 hour, and the secondary antibodies were incubated at a concentration equal to that of each primary antibody for 5 days at 37 °C. Wash the samples three times for 1 hour at room temperature with a wash buffer to remove excess antibodies. Dehydrate gradually in EtOH/H2O (50% ×1, 70% ×1, 80% ×1, 100% ×2, 6 h for each step). Switch to DCM for 1 h and then to BABB until the tissues become transparent. Finally, the results will be examined with a light-sheet microscope for volume imaging.
Image stacks were acquired using a Blaze ultramicroscope or II ultramicroscope (LaVision BioTec) with an axial resolution of 4 μm and the following filter sets: ex 470/40 nm, em 535/50 nm; ex 545/25 nm, em 605/70 nm; ex 640/40 nm, em 690/50 nm. Entire mouse bodies were scanned individually with a low magnification Ultramicroscope Blaze light sheet microscopy objective: 1.1× objective (LaVision BioTec MI PLAN 1.1×/0.1 NA [WD = 17 mm]). We covered the entire mouse with 3 × 8 tile scans with 25% overlap and imaged from the ventral and dorsal surfaces to 11 mm in depth, covering the entire body volume with a Z-step of 10 μm. The width of the light-sheet was kept at 100%, and the exposure time was set at 120 ms. The laser power was adjusted depending on the intensity of the fluorescence signal to avoid saturation. High-magnification tile scans for multiple organs (including brain, heart, lung, thymus, liver, spinal cord, spleen, kidney, intestine, testis, and ovary) were acquired individually with high-magnification objectives (Olympus XLFLUOR 4× corrected/0.28 NA [WD = 10 mm] and LaVision BioTec MI PLAN 12×/0.53 NA [WD = 10 mm]) coupled to an Olympus rotary zoom unit (U-TVCAC) set at 1×. The high-magnification tile scans were acquired with a 20% overlap, and the light-sheet width was reduced to achieve maximum illumination in the field of view. The acquired raw images TIFF were processed with Fiji’s stitching plugin, then with Vision4D (v.3.3 × 64, Arivis) for volume fusion, and visualized in Imaris (v.9.6 × 64, Imaris) for 3D reconstruction, analysis, and video generation.
Briefly, 10 μm thick frozen sections were treated with 0.2% Triton X-100 in PBS for 15 minutes and blocked with 10% serum in PBST for 40 minutes at room temperature. They were then incubated with primary antibodies, SARS-CoV-2 (COVID-19) Spike (1:500, GeneTex, GTX135356, GTX632604), Nucleocapsid (1:500, Invitrogen, PA1-41098), ACE2 (1:500, Invitrogen, PA5-20039), NeuN (1:500, Invitrogen, PA5-78499), c-caspase3 (1:500, Cell Signaling Technology, 9664), APP (1:500, Invitrogen, 51-2700), F4/80 (1:500, Abcam, ab6640), Anti-Influenza A H1N1 hemagglutinin antibody (1:500, Abcam, ab128412), ATPase antibody (1:500, Abcam, ab76020), CD31 (1:500, Abcam, ab7388), AQP4 (1:500, Abcam, ab9512), PDGFR (1:500, Abcam, ab32570) overnight at 4°C and washed with PBS for 15 min, Alexa-conjugated secondary antibodies (1:1000, goat anti-rabbit IgG Alexa Fluor 647, Invitrogen, A21245; goat anti-rabbit IgG Alexa Fluor 568, Invitrogen, A-11036; goat anti-mouse IgG Alexa Fluor 568, Invitrogen, A-11031; goat anti-mouse IgG Alexa Fluor 647, Invitrogen, A-21235; goat anti-rat IgG Alexa Fluor 568, Invitrogen, A-11077) were incubated for 1 hour at room temperature. Sections were mounted after staining with Hoechst 33342 (Invitrogen). Images of a confocal microscope were acquired with a 40x immersion objective (Zeiss, EC Plan-Neofluar 40x/1.30 Oil DIC M27) (ZEISS LSM880). Three random regions (5mm x 5mm x 5mm) were dissected to detect spike protein in each sample, and 20 sections were prepared from each region for immunostaining.
Briefly, frozen sections were permeabilized in 0.1% Triton X-100 in PBS, treated with 3% H2O2 for 10 min, and blocked with 10% goat serum for 20 min, incubated with primary antibody against SARS-CoV-2 (COVID-19) Spike (1:200, GeneTex, GTX135356) at room temperature for 2 h. Staining was detected with goat anti-rabbit IgG HRP antibody (1:200, Abcam, ab6721) and revealed by incubation with diaminobenzidine for 10-20 seconds (Vectorlabs, VEC-MP-7714). Hematoxylin (Sigma, 51275) was used as counterstaining. Prussian blue staining was performed according to the manufacturer’s instructions (NovaUltra, IW-3010).
Decalcified skull samples were minced in PBS, meninges and brain samples were ground in liquid nitrogen and dissolved in PBS, and the tissue extract was passed through 40 μm strainer. RNA extraction was performed using RNeasy FFPE Kit (QIAGEN, 73504), followed by SARS-CoV-2 quantitative reverse-transcription PCR using the Seegene Allplex™ 2019-nCoV Assay (cat. no: RP10243X) to probe for the E gene, RdRp/S gene, and N gene on a CFX96 Real-time PCR Detection System-IVD (Bio-Rad). Samples with Ct values below 38 were deemed positive.
We collected CSF samples from 35 COVID-19 patients with moderate to severe symptoms. Among these, 27 patients provided CSF samples three months post-infection, while eight samples were obtained during the acute phase, within one month of infection. We classified eleven individuals with neurological symptoms three months post-infection as Long COVID patients, according to WHO criteria, and included 20 non-COVID controls without neurological diseases (Table S3). CSF levels of total tau, NfL, GFAP, and UCHL1 were measured using a Neurology 4-Plex A assay kit (Quanterix, Billerica, MA, USA) and SIMOA technology (Quanterix, Billerica, MA, USA) following the manufacturer’s instructions.
For microdissection of the spike protein-positive regions, we laser-cut and isolated the selected samples using the laser capture system (Leica, LMD7000). Briefly, the cryosections of the human brain were mounted on a Polyethylene naphthalate (PEN) Membrane Slide (Zeiss, 415190-9041-000) and then stained with spike protein antibodies using the IHC kit (Abcam, ab210062). After further processing, the sections were serially dehydrated with ethanol and air-dried under a fume hood for 15 minutes. The spike protein-positive and spike protein-negative regions of the COVID-19 human brain sections were selected with a closed-shape manual drawing tool and dissected using a UV laser. The excised regions were collected into a 0.5 ml tube and examined by camera. We cut and pooled 30 regions as one sample; an accumulated area of 6 mm2 was collected by laser cut and using 40× objective (HC PL FL L 40×/0.60 XT CORR). The tissues were quickly spun down and stored at -80 °C for further proteomic analysis.
Sample preparation for proteomics analysis was performed as described previously with slight modifications.61 The human samples consisted of post-mortem skulls with meninges tissues from ten COVID-19 deceased and ten control donors and post-mortem brain cortex tissues from eleven COVID-19 deceased and eight control donors. The mouse samples consisted of skull, meninges, and brain cortex tissues from WT mice injected with spike S1, HA, and saline control, four mice in each group. Protein extraction was carried out from 4% neutral buffered formalin-fixed tissues. Briefly, the cells from the skull and the meninges tissues were isolated by mincing or grinding, and the tissue extract was passed through a 40 μm strainer. The cell pellet was washed with PBS and resuspended in SDS-lysis buffer (6% Sodium dodecyl sulfate, 500 mM TrisHCl, pH 8.5). The brain samples were pulverized in a Covaris CPO2 before adding an SDS-lysis buffer. This was followed by heating at 95°C for 45 min at 1000 rpm in a thermomixer. The samples were then subjected to ultrasonication using a Bioruptor Pico sonication device operated at high frequency for 30 sec on and off for 30 cycles. After ultrasonication, the samples were again heated at 95°C for 45 min at 1000 rpm in a thermomixer. This was followed by protein precipitation in ice-cold acetone (80% v/v) overnight at -80°C, followed by centrifugation for 15 min at 4°C. For reduction and alkylation, the proteins were resuspended in SDC buffer and heated at 95°C for 10 min with 1000 rpm. Trypsin and LysC digestion were carried out at an enzyme/substrate ratio of 1:50, and the samples were incubated at 37°C overnight at 1000 rpm in a thermomixer. Next, peptides were acidified using 1% TFA in 99% isopropanol in a 1:1 v/v ratio. The peptides were subjected to in-house built StageTips consisting of two layers of styrene-divinylbenzene reversed-phase sulfonate (SDB-RPS; 3 M Empore) membranes. Peptides were loaded on the activated (100% ACN, 1% TFA in 30% Methanol, 0.2% TFA, respectively) StageTips, run through the SDB-RPS membranes, and washed by EtOAc including 1% TFA, isopropanol including 1% TFA, and 0.2% TFA, respectively. Peptide elution was carried out in 60 μL of 1.25% Ammonia, 80% ACN and dried for 40 min at 45 °C in a SpeedVac (Eppendorf, Concentrator plus). The dried peptides were reconstituted in 10 μL of 2% ACN/0.1% TFA and peptide concentration was estimated using Pierce™ Quantitative Colorimetric Peptide Assay.
The mass spectrometry data was generated through data-independent acquisition (DIA) modes. The LC-MS/MS analysis was carried out using EASY nanoLC 1200 (Thermo Fisher Scientific) coupled with trapped ion mobility spectrometry quadrupole time-of-flight single cell proteomics mass spectrometer (timsTOF SCP, Bruker Daltonik GmbH, Germany) via a CaptiveSpray nano-electrospray ion source. Peptides (50 ng) were loaded onto a 25 cm Aurora Series UHPLC column with CaptiveSpray insert (75 μm ID, 1.6 μm C18) at 50°C and separated using a 50 min gradient (5-20% buffer B in 30 min, 20-29% buffer B in 9 min, 29-45% in 6 min, 45-95% in 5 min, wash with 95% buffer B for 5 min, 95-5% buffer B in 5 min) at a flow rate of 300 nL/min. Buffer A and B were water with 0.1 vol% formic acid and 80:20:0.1 vol% ACN:water:formic acid, respectively. MS data were acquired in single-shot library-free DIA mode and the timsTOF SCP was operated in DIA/parallel accumulation serial fragmentation (PASEF) using the high sensitivity detection-low sample amount mode. The ion accumulation and ramp time was set to 100 ms each to achieve nearly 100% duty cycle. The collision energy was ramped linearly as a function of the mobility from 59 eV at 1/K0 = 1.6 V-s cm−2 to 20 eV at 1/K0 = 0.6 V-s cm−2. The isolation windows were defined as 24 × 25 Th from m/z 400 to 1000.
Young neonatal mouse pups, aged 7-9 days postnatal, were decapitated in compliance with animal handling regulations. Brains were carefully removed, and the hippocampi and neocortices were dissected. We cut 350 μm thick sagittal sections using a Mcllwain tissue chopper (Model TC752, Mickle Laboratory Engineering Company) and selected intact sections under a dissection microscope (SZ61, Olympus). These sections were subjected to a 30 min incubation at 4°C in pre-cooled dissection media consisting of 50% HEPES-buffered MEM (32360-026, Gibco), 1% penicillin–streptomycin, and 10 mM Tris, pH 7.2. Subsequently, we plated two slices onto each 0.4-μm porous polytetrafluoroethylene (PTFE) membrane insert (PICM ORG 50, Millipore) within a 3.5- cm dish filled with 1 ml of slice culture media. The slice culture media consisted of 50% HEPES-buffered MEM (32360-026, Gibco), 25% heat-inactivated horse serum (26050-088, Gibco), 25% HBSS (14025-050, Gibco), and 1 mM L-glutamine (M11-004, Gibco) at pH 7.4. The cultures were maintained in a cell-culture incubator at 37°C, 5% CO2. Media exchange was performed on the first day following preparation and subsequently every two days to maintain optimal conditions for the slices. After 14 days, the brain sections were treated with a mixture solution of 990 μl media and 10 μl 1 mg/ml spike protein, denatured spike protein or PBS control. 1 mg/ml spike protein stock solution was heated at 95°C for 30 min for denaturation. Regarding pre-treatment with ACE2 antibody, 10 μl ACE2 antibody solution was added to 990 μl media for 1 hour before changing to spike protein or PBS control. At 3 days after spike protein treatment, the culture media were collected for ELISA, and brain slices were fixed in 4% paraformaldehyde (PFA) and sucrose solution at pH 7.4 for 15 minutes at room temperature (RT), followed by three PBS washes. Fixed slices were permeabilized for 30 minutes in 1XPBS (10010023, Gibco), containing 0.5% Triton X-100. Slices were delicately excised from the membrane inserts using a scalpel and then transferred to a wet chamber. Subsequently, they were subjected to a 1-hour blocking step in a solution of PBS with 0.5% Triton X-100 and 5% goat serum (16210064, Life) at RT. The slices were then incubated overnight at 4°C in the blocking solution with primary antibodies. After the primary antibody incubation, the slices were washed three times for 10 minutes each with PBS containing 0.5% Triton X-100. Subsequently, slices were incubated for 3-5 hours at RT in the blocking solution containing Hoechst 3342 (H1399, Invitrogen, 1:2000) and appropriate secondary antibodies conjugated to Alexa Fluor 488, 555, and 647 (all Life technologies, 1:500). Following the secondary antibody incubation, slices underwent three additional 10-minutes washes. Finally, slices were mounted using Gel Mount Aqueous Mounting Medium (G0918, Sigma-Aldrich) and analyzed using confocal microscopy.
At 1 hour after spike protein treatment, brain slices were lysed in ice-cold RIPA buffer (50mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA and 1% Triton X-100) and incubated with 0.1% SDS on ice for 10 mins and centrifuged to remove debris. Protein concentration was measured using a BCA assay (Thermo Fisher Scientific, 10741395). Protein lysates were then mixed with 4x LDS Sample Buffer (NuPAGE™ Thermo Fisher Scientific, NP0008) containing 2-mercaptoethanol (Sigma-Aldrich, M6250) and incubated at 70°C for 10 min. Samples were run on a 10% SDS-PAGE gel. EZ-Run™ Prestained Rec Protein Ladder (Fisher BioReagents™, BP3603500) was used to assess the molecular weights. Proteins were transferred to 0.2 μm PVDF membranes (Merck Millipore, ISEQ00010) and blocked with 5% skim milk (Biomol, S1013-90A.500) in 10 mM Tris base saline -0.1% Tween 20 (TBST) for 1 h at room temperature. Primary antibodies were diluted with Starting blocking buffer (Thermo Scientific, 37542), and HRP conjugated secondary antibodies were diluted in 5% skim milk in TBST. Amersham Hyperfilm ECL (GE, GE28-9068-36) and HRP substrate ECL (Merck Millipore, WBKLS0500) were used to detect signals. Band intensities were quantified using ImageLab (Bio-Rad version 6.1.0). The following primary and secondary antibodies were used for blotting.
Mouse blood was collected into citrate-treated tubes and centrifuged at 2000 x g for 15 minutes, and the supernatant was immediately subjected to the ELISA assay according to the manufacturer’s protocol. Mouse Interferon Gamma (IFNg) ELISA Kit (RD-IFNg-Mu, Reddot biotech), Mouse Interleukin 6 (IL-6) ELISA Kit (RD-IL6-Mu, Reddot biotech), Mouse Interferon Gamma Induced Protein 10kDa (IP10) ELISA Kit (RD-IP10-Mu, Reddot biotech) were used in this study.
The experimental setup included a gray open field measuring 60 cm by 60 cm by 26 cm and overhead cameras coupled with the computer tracking system (Noldus EthoVision XT). The mice underwent handling for one minute daily and were introduced to the apparatus for 5 minutes for habituation over two days, and the arena was cleaned with ethanol between mice. On the day of open field tests, the mouse was introduced into the arena, allowing it to explore freely for 20 minutes. The movements were captured on video, and the total distance it traveled and the time it spent in different areas were quantified. In the training phase of novel object recognition tests, the mice were placed in the apparatus with two identical objects, where they were allowed to explore for 5 minutes. After a 24-hour retention period, the mice were reintroduced to the apparatus, this time with one of the original objects replaced by a novel object with a different shape and color but similar height, and were again allowed to explore for 5 minutes. The objects used in the experiment were plastic rectangular boxes.
The MCAo model induced transient cerebral ischemic strokes in mice under isoflurane anesthesia (30% O2, 70% N2O). We ligated the left common and internal carotid arteries, inserted a silicon-capped nylon suture (6/0) through a cut in the common carotid artery, and pushed it further into MCA, occluding the MCA for 30 minutes. Cerebral blood flow (2 mm posterior, 5 mm lateral from bregma) was monitored with a transcranial laser Doppler flowmeter. After reperfusion, mice recovered for two hours in temperature-controlled cages. Sham-operated mice underwent a similar procedure without filament insertion. Consistent body temperatures (37.0 ± 0.5°C) were maintained with a heating pad. After surgery, mice were given mashed food in a petri dish on the floor to promote eating. We excluded mice with insufficient MCA occlusion (<15% reduction in blood flow) or >80% blood flow recovery within 10 minutes of reperfusion.
After 4 weeks of spike protein or saline injection, the animals were subjected to traumatic brain injury by controlled cortical impact (CCI) as described before.110 Briefly, 30 minutes after an injection of buprenorphine (0.1mg/kg, single shot ip), animals were anesthetized with 5% isoflurane in a gas mixture of 30% oxygen and 70% nitrogen for 90 seconds. Anesthesia was maintained by an isoflurane application of 1.5-2% afterward. A feedback-controlled heating pad kept the body temperature at 37°C. The head of the mouse was fixed in a stereotaxic frame, and a rectangular craniotomy (4×4 mm) was prepared posterior-lateral to bregma over the right parietal cortex under continuous cooling with saline, leaving the dura mater intact. Subsequently, CCI was induced by a custom-made device (L. Kopacz, University of Mainz, Germany; diameter: 3.0 mm, velocity 6 m/sec, penetration depth 0.5 mm, contact time of 150 ms). The craniotomy was resealed with the previously removed bone flap, the skin incision was sutured, and animals were placed in an incubator warmed at 37°C for recovery. Magnetic resonance imaging (MRI) and data analysis were performed as described.111 Mice were anesthetized with isoflurane (5% for induction and 1-1.5% for maintenance of anesthesia in a gas mic of 30% oxygen and 70% nitrogen), and body temperature, heart rate, and respiration rate were continuously monitored. MRI scans were performed on a 3T nanoScan® PET/MR (Mediso, Münster, Germany), including 3D T1-weighted and 2D T2-weighted sequences. Each imaging session lasted around 20 min and was performed at 4 h, 24 h, 72 h, 7 days, 14 days, 28 days, and 42 days post CCI operation. Lesion volume and hemispheric swelling were determined based on T2-weighted sequences by using ImageJ (https://imagej.nih.gov/ij/). Briefly, we selected 14 MRI slices equidistant to each other, covering the lesion area. The lesion and the area of each hemisphere were quantified (A), and lesion volume and hemispheric volume (V) were then calculated using the following formula: V= d∗(A1/2 + A2 + A3…+ An/2), with d being the distance between slices in millimeter. Hemispheric swelling was calculated as the ratio of ipsilateral to contralateral hemispheric volume. Segmentation and measurement of the volume of the lateral and third ventricles were performed using ITK-SNAP software112 on the T2w sections. Then, net brain tissue volumes on the contralateral side were calculated by subtracting the ventricular volume from the corresponding hemispheric volume. On the ipsilateral side, the volume of net brain tissue was assessed by subtracting the ventricle’s and the lesion’s volume from the hemispheric volume. Finally, ipsilateral brain tissue loss was determined by subtracting the volume of net brain tissue on the ipsilateral side from the contralateral side.
Up to 30mg of lung tissue was transferred into 2ml reinforced screw cap tubes (VWR, Radnor, PA, USA) containing 2.8mm ceramic beads (VWR, Radnor, PA, USA) and 500ml of RNAlater (Qiagen, Hilden, Germany). Samples were stored until processing at -80°C. For RNA isolation, RNAlater was replaced by 600ml of RA1 buffer of the NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany) containing ß-mercaptoethanol (Sigma-Aldrich, Burlington, MA, USA). Tissue was homogenized with the Precellys Evolution (Bertin-Technologies, Montigny-le-Bretonneux, France) applying two crushing cycles of 20 sec at 5000 rpm with a cooling phase between cycles of 2 min at 4°C. RNA was extracted according to the manufacturer’s protocol. RNA was quantified using a Nanodrop (Thermo Fisher, Waltham, MA, USA), and cDNA synthesis was performed with the Takara Prime Script RT Master Mix (RR036A, Takara Bio, San Jose, CA, USA) according to the manufacturer’s instructions. Absolute quantitative real-time PCR was performed using PowerTrack SYBR Green Master Mix using a QuantStudio 5 (Thermo Fisher, Waltham, MA, USA) and a SARS-CoV-2 encoding plasmid standard with a serial dilution of 109 – 101 copies. 1mM final concentration of forward primer (GACCCCAAAATCAGCGAAAT) and reverse primer (TCTGGTTACTGCCAGTTGAATCTG) against CDC-N1 (2019-nCoV_N1) were used applying the following thermocycler amplification parameters for 45 cycles: 95°C for 15 sec, 55°C for 10 sec and 72° for 25 sec. Data analysis was performed using the system-integrated DA1 software.
diaPASEF raw files were searched against the human and mouse Uniprot database using DIA-NN.113 A peptide length range of seven amino acids was considered for the search, including N-terminal acetylation. Methionine oxidation was set as variable modifications and cysteine carbamidomethylation as fixed modification. Enzyme specificity was set to Trypsin/P with 2 missed cleavages. The FASTA digest for the library-free search was enabled to predict library generation. The FDR was set to 1% at precursor and global protein levels. Match-between-runs (MBR) feature was enabled, and the quantification mode was set to “Robust LC (high precision).” The Protein Group column in DIA-NN’s report was used to identify the protein group and PG.MaxLFQ was used to calculate the differential expression.
Proteomics data analysis was performed in Perseus and R studio. The protein groups were filtered so that only those proteins were considered for differential expression, which was present in 70% of samples in each group with valid values. The values were log2 transformed and normalized with the median centering of the dataset. The missing values were randomly drawn from a Gaussian distribution with a width of 0.3 standard deviations downshifted by 1.8 standard deviations. The correlation heatmap was computed using the Pearson correlation coefficient. Differential protein expression between the COVID-19 and control groups was performed using a student-t test. The Benjamini-Hochberg procedure was applied to correct for multiple comparisons. P-value < 0.05 and fold change > 1.5 was taken as statistical significance unless specified. The gene ontology (GO) and pathway enrichment were carried out in Cytoscape using ClueGO plug-in114 or ClusterProfiler. PCA plot, volcano plot, GO_Chord plot, and heatmap were generated in R studio.
Prism 8.0 was used for all statistical calculations (GraphPad Software, San Diego, CA). The number of independent biological replicates and statistical tests used are specified in all figure legends. The data were analyzed using the Shapiro-Wilk normality test with Gaussian distribution.
Paterson, R.W. ∙ Brown, R.L. ∙ Benjamin, L. …
The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings
Brain. 2020; 143:3104-3120
Tu, T.M. ∙ Seet, C.Y.H. ∙ Koh, J.S. …
Acute Ischemic Stroke During the Convalescent Phase of Asymptomatic COVID-2019 Infection in Men
JAMA Netw. Open. 2021; 4, e217498
Meinhardt, J. ∙ Radke, J. ∙ Dittmayer, C. …
Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19
Nat. Neurosci. 2021; 24:168-175
Krasemann, S. ∙ Dittmayer, C. ∙ von Stillfried, S. …
Assessing and improving the validity of COVID-19 autopsy studies – A multicentre approach to establish essential standards for immunohistochemical and ultrastructural analyses
EBioMedicine. 2022; 83, 104193
Fullard, J.F. ∙ Lee, H.-C. ∙ Voloudakis, G. …
Single-nucleus transcriptome analysis of human brain immune response in patients with severe COVID-19
Genome Med. 2021; 13:118
Frank, M.G. ∙ Nguyen, K.H. ∙ Ball, J.B. …
SARS-CoV-2 spike S1 subunit induces neuroinflammatory, microglial and behavioral sickness responses: Evidence of PAMP-like properties
Brain Behav. Immun. 2022; 100:267-277
Raghavan, S. ∙ Kenchappa, D.B. ∙ Leo, M.D.
SARS-CoV-2 Spike Protein Induces Degradation of Junctional Proteins That Maintain Endothelial Barrier Integrity
Front. Cardiovasc. Med. 2021; 8, 687783
Kim, E.S. ∙ Jeon, M.-T. ∙ Kim, K.-S. …
Spike Proteins of SARS-CoV-2 Induce Pathological Changes in Molecular Delivery and Metabolic Function in the Brain Endothelial Cells
Viruses. 2021; 13, 2021
DeOre, B.J. ∙ Tran, K.A. ∙ Andrews, A.M. …
SARS-CoV-2 Spike Protein Disrupts Blood-Brain Barrier Integrity via RhoA Activation
J. Neuroimmune Pharmacol. 2021; 16:722-728
Nuovo, G.J. ∙ Magro, C. ∙ Shaffer, T. …
Endothelial cell damage is the central part of COVID-19 and a mouse model induced by injection of the S1 subunit of the spike protein
Ann. Diagn. Pathol. 2021; 51, 151682
Khan, S. ∙ Shafiei, M.S. ∙ Longoria, C. …
SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway
eLife. 2021; 10, e68563
Patterson, B.K. ∙ Francisco, E.B. ∙ Yogendra, R. …
Persistence of SARS CoV-2 S1 Protein in CD16+ Monocytes in Post-Acute Sequelae of COVID-19 (PASC) up to 15 Months Post-Infection
Front. Immunol. 2021; 12, 746021
Swank, Z. ∙ Senussi, Y. ∙ Manickas-Hill, Z. …
Persistent Circulating Severe Acute Respiratory Syndrome Coronavirus 2 Spike Is Associated With Post-acute Coronavirus Disease 2019 Sequelae
Clin. Infect. Dis. 2023; 76:e487-e490
Butowt, R. ∙ Meunier, N. ∙ Bryche, B. …
The olfactory nerve is not a likely route to brain infection in COVID-19: a critical review of data from humans and animal models
Acta Neuropathol. 2021; 141:809-822
Cai, R. ∙ Pan, C. ∙ Ghasemigharagoz, A. …
Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull–meninges connections
Nat. Neurosci. 2019; 22:317-327
Herisson, F. ∙ Frodermann, V. ∙ Courties, G. …
Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration
Nat. Neurosci. 2018; 21:1209-1217
Cugurra, A. ∙ Mamuladze, T. ∙ Rustenhoven, J. …
Skull and vertebral bone marrow are myeloid cell reservoirs for the meninges and CNS parenchyma
Science. 2021; 373, eabf7844
Brioschi, S. ∙ Wang, W.-L. ∙ Peng, V. …
Heterogeneity of meningeal B cells reveals a lymphopoietic niche at the CNS borders
Science. 2021; 373, eabf9277
Schafflick, D. ∙ Wolbert, J. ∙ Heming, M. …
Single-cell profiling of CNS border compartment leukocytes reveals that B cells and their progenitors reside in non-diseased meninges
Nat. Neurosci. 2021; 24:1225-1234
A local source of immune cells for the central nervous system
Published online June 17, 2021Nat. Med. 2021;
Leasure, A.C. ∙ Khan, Y.M. ∙ Iyer, R. …
Intracerebral Hemorrhage in Patients With COVID-19: an Analysis From the COVID-19 Cardiovascular Disease Registry
Stroke. 2021; 52:e321-e323
Niu, Z. ∙ Zhang, Z. ∙ Gao, X. …
N501Y mutation imparts cross-species transmission of SARS-CoV-2 to mice by enhancing receptor binding
Signal Transduct. Target. Ther. 2021; 6:284
Hamming, I. ∙ Timens, W. ∙ Bulthuis, M.L. …
Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis
J. Pathol. 2004; 203:631-637
Beyerstedt, S. ∙ Casaro, E.B. ∙ Rangel, É.B.
COVID-19: angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection
Eur. J. Clin. Microbiol. Infect. Dis. 2021; 40:905-919
Zhang, Y. ∙ Archie, S.R. ∙ Ghanwatkar, Y. …
Potential role of astrocyte angiotensin converting enzyme 2 in the neural transmission of COVID-19 and a neuroinflammatory state induced by smoking and vaping
Fluids Barriers CNS. 2022; 19:46
Pérez-Mies, B. ∙ Caniego-Casas, T. ∙ Bardi, T. …
Progression to lung fibrosis in severe COVID-19 patients: A morphological and transcriptomic study in postmortem samples
Front. Med. (Lausanne). 2022; 9, 976759
Zhou, Y.-Q. ∙ Wang, K. ∙ Wang, X.-Y. …
SARS-CoV-2 Pseudovirus enters the host cells through spike protein-CD147 in an Arf6-dependent manner
Emerg. Microbes Infect. 2022; 11:1135-1144
Bonetto, V. ∙ Pasetto, L. ∙ Lisi, I. …
Markers of blood-brain barrier disruption increase early and persistently in COVID-19 patients with neurological manifestations
Front. Immunol. 2022; 13, 1070379
Geisheker, M.R. ∙ Heymann, G. ∙ Wang, T. …
Hotspots of missense mutation identify neurodevelopmental disorder genes and functional domains
Nat. Neurosci. 2017; 20:1043-1051
Ang, G. ∙ Brown, L.A. ∙ Tam, S.K.E. …
Deletion of AMPA receptor GluA1 subunit gene (Gria1) causes circadian rhythm disruption and aberrant responses to environmental cues
Transl. Psychiatry. 2021; 11:588
Zhao, S. ∙ Shibata, K. ∙ Hellyer, P.J. …
Rapid vigilance and episodic memory decrements in COVID-19 survivors
Brain Commun. 2022; 4, fcab295
Simpson, J.E. ∙ Ince, P.G. ∙ Lace, G. …, MRC Cognitive Function and Ageing Neuropathology Study Group
Astrocyte phenotype in relation to Alzheimer-type pathology in the ageing brain
Neurobiol. Aging. 2010; 31:578-590
Deloulme, J.-C. ∙ Gory-Fauré, S. ∙ Mauconduit, F. …
Microtubule-associated protein 6 mediates neuronal connectivity through Semaphorin 3E-dependent signalling for axonal growth
Nat. Commun. 2015; 6, 7246
Gil, O.D. ∙ Zanazzi, G. ∙ Struyk, A.F. …
Neurotrimin Mediates Bifunctional Effects on Neurite Outgrowth via Homophilic and Heterophilic Interactions
J. Neurosci. 1998; 18:9312-9325
Scheper, W. ∙ Zwart, R. ∙ Baas, F.
Rab6 membrane association is dependent of Presenilin 1 and cellular phosphorylation events
Brain Res. Mol. Brain Res. 2004; 122:17-23
Shi, X. ∙ Ohta, Y. ∙ Liu, X. …
Acute Anti-Inflammatory Markers ITIH4 and AHSG in Mice Brain of a Novel Alzheimer’s Disease Model
J. Alzheimers. Dis. 2019; 68:1667-1675
Gentleman, S.M. ∙ Nash, M.J. ∙ Sweeting, C.J. …
β-Amyloid precursor protein (βAPP) as a marker for axonal injury after head injury
Neurosci. Lett. 1993; 160:139-144
Wright, G.J. ∙ Puklavec, M.J. ∙ Willis, A.C. …
Lymphoid/neuronal cell surface OX2 glycoprotein recognizes a novel receptor on macrophages implicated in the control of their function
Immunity. 2000; 13:233-242
Behesti, H. ∙ Fore, T.R. ∙ Wu, P. …
ASTN2 modulates synaptic strength by trafficking and degradation of surface proteins
Proc. Natl. Acad. Sci. USA. 2018; 115:E9717-E9726
Dejanovic, B. ∙ Semtner, M. ∙ Ebert, S. …
Palmitoylation of Gephyrin Controls Receptor Clustering and Plasticity of GABAergic Synapses
PLoS Biol. 2014; 12, e1001908
Xing, G. ∙ Ren, M. ∙ O’Neill, J.T. …
Controlled cortical impact injury and craniotomy result in divergent alterations of pyruvate metabolizing enzymes in rat brain
Exp. Neurol. 2012; 234:31-38
Tamura, T. ∙ Irie, T. ∙ Deguchi, S. …
Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant
Nat. Commun. 2024; 15, 1176
Vanderheiden, A. ∙ Hill, J.D. ∙ Jiang, X. …
Vaccination reduces central nervous system IL-1β and memory deficits after COVID-19 in mice
Nat. Immunol. 2024; 25:1158-1171
Greene, C. ∙ Connolly, R. ∙ Brennan, D. …
Blood–brain barrier disruption and sustained systemic inflammation in individuals with long COVID-associated cognitive impairment
Nat. Neurosci. 2024; 27:421-432
Gold, J.E. ∙ Okyay, R.A. ∙ Licht, W.E. …
Investigation of Long COVID Prevalence and Its Relationship to Epstein-Barr Virus Reactivation
Pathogens. 2021; 10:763
Letarov, A.V. ∙ Babenko, V.V. ∙ Kulikov, E.E.
Free SARS-CoV-2 Spike Protein S1 Particles May Play a Role in the Pathogenesis of COVID-19 Infection
Biochemistry (Mosc). 2021; 86:257-261
Oh, J. ∙ Cho, W.-H. ∙ Barcelon, E. …
SARS-CoV-2 spike protein induces cognitive deficit and anxiety-like behavior in mouse via non-cell autonomous hippocampal neuronal death
Sci. Rep. 2022; 12:5496
Zhang, L. ∙ Zhou, L. ∙ Bao, L. …
SARS-CoV-2 crosses the blood–brain barrier accompanied with basement membrane disruption without tight junctions alteration
Signal Transduct. Target. Ther. 2021; 6:337
Maugeri, N. ∙ Campana, L. ∙ Gavina, M. …
Activated platelets present high mobility group box 1 to neutrophils, inducing autophagy and promoting the extrusion of neutrophil extracellular traps
J. Thromb. Haemost. 2014; 12:2074-2088
Pramitasuri, T.I. ∙ Laksmidewi, A.A.A.P. ∙ Putra, I.B.K. …
Neutrophil Extracellular Traps in Coronavirus Disease-19-Associated Ischemic Stroke: A Novel Avenue in Neuroscience
Exp. Neurobiol. 2021; 30:1-12
Veras, F.P. ∙ Pontelli, M.C. ∙ Silva, C.M. …
SARS-CoV-2–triggered neutrophil extracellular traps mediate COVID-19 pathology
J. Exp. Med. 2020; 217, e20201129
Middleton, E.A. ∙ He, X.-Y. ∙ Denorme, F. …
Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome
Blood. 2020; 136:1169-1179
Veras, F.P. ∙ Gomes, G.F. ∙ Silva, B.M.S. …
Targeting neutrophils extracellular traps (NETs) reduces multiple organ injury in a COVID-19 mouse model
Respir. Res. 2023; 24:66
Qiu, Y. ∙ Wu, D. ∙ Ning, W. …
Post-mortem tissue proteomics reveals the pathogenesis of multi-organ injuries of COVID-19
Natl. Sci. Rev. 2021; 8, nwab143
Zweckberger, K. ∙ Stoffel, M. ∙ Baethmann, A. …
Effect of decompression craniotomy on increase of contusion volume and functional outcome after controlled cortical impact in mice
J. Neurotrauma. 2003; 20:1307-1314
Hu, S. ∙ Exner, C. ∙ Sienel, R.I. …
Characterization of Vasogenic and Cytotoxic Brain Edema Formation After Experimental Traumatic Brain Injury by Free Water Diffusion Magnetic Resonance Imaging
J. Neurotrauma. 2024; 41:393-406
Yushkevich, P.A. ∙ Piven, J. ∙ Hazlett, H.C. …
User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability
NeuroImage. 2006; 31:1116-1128
Demichev, V. ∙ Messner, C.B. ∙ Vernardis, S.I. …
DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput
Nat. Methods. 2020; 17:41-44
Bindea, G. ∙ Mlecnik, B. ∙ Hackl, H. …
ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks
Bioinformatics. 2009; 25:1091-1093